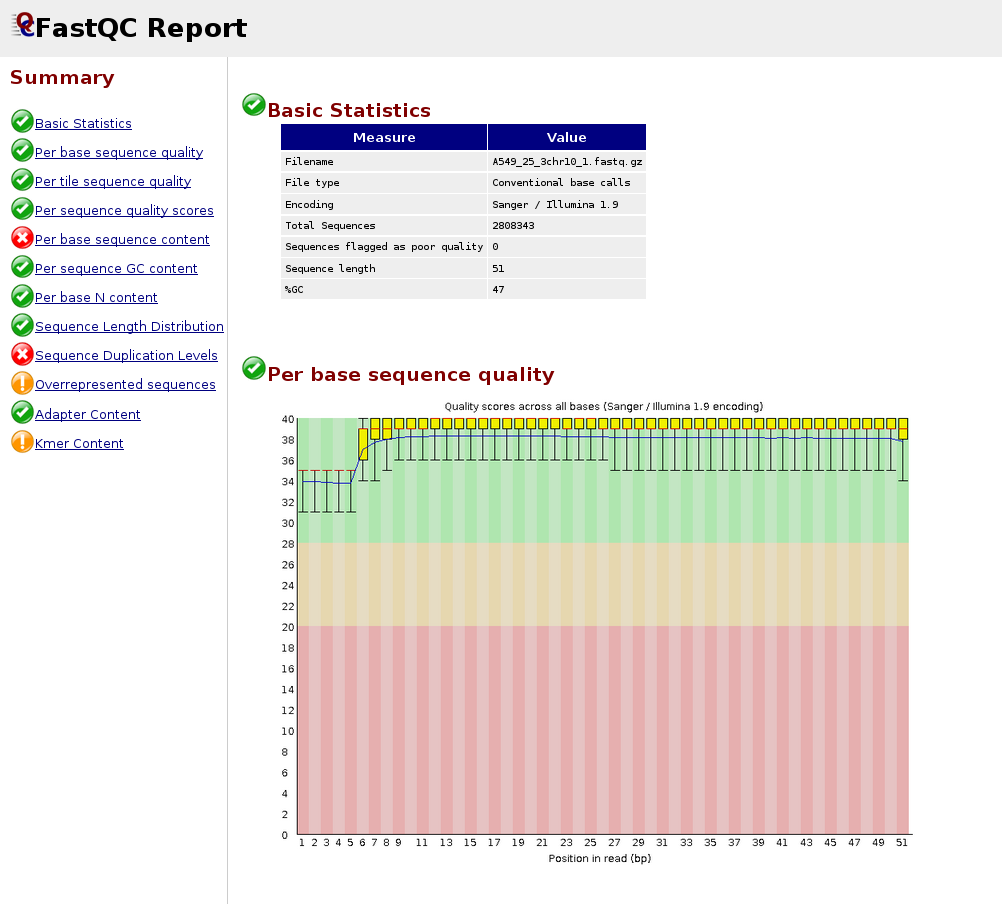
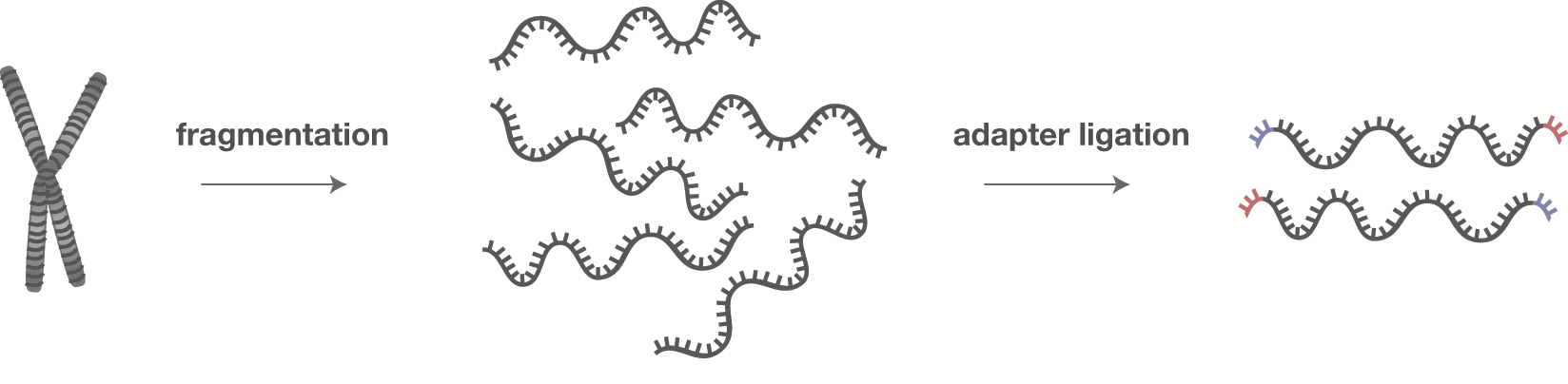
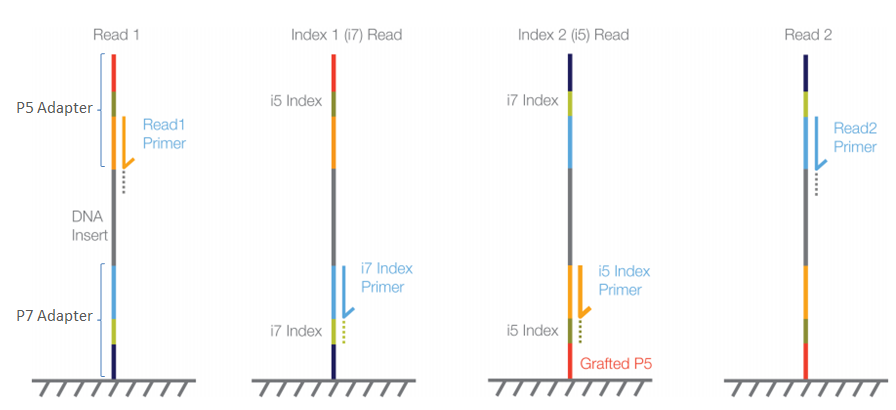
Adapter trimming for single end sRNA-seq data. The reads processed by... | Download Scientific Diagram
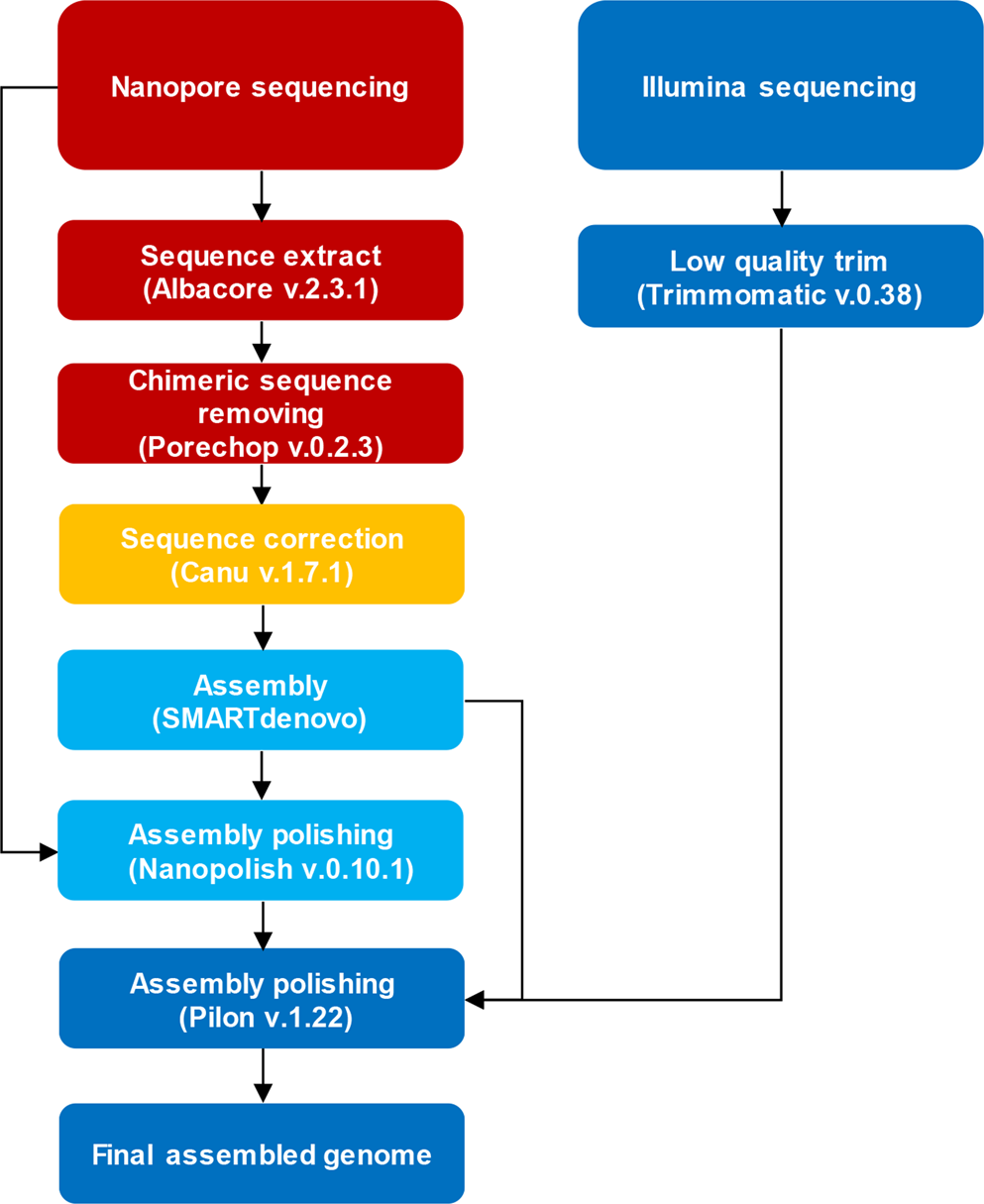
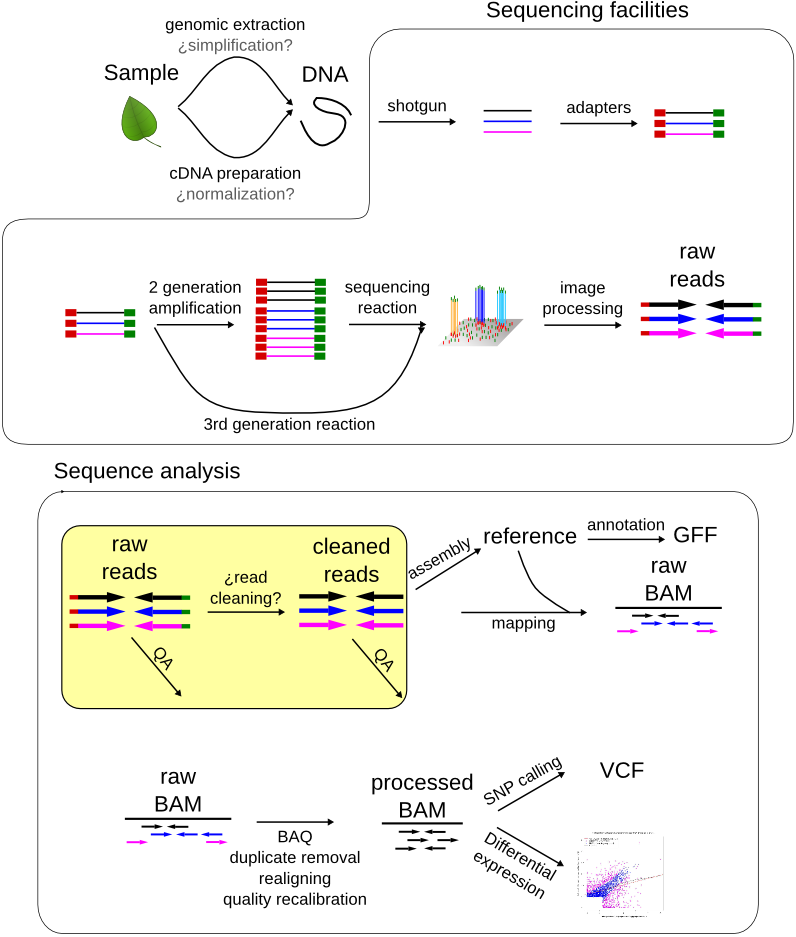
Nanopore sequencing reads improve assembly and gene annotation of the Parochlus steinenii genome | Scientific Reports
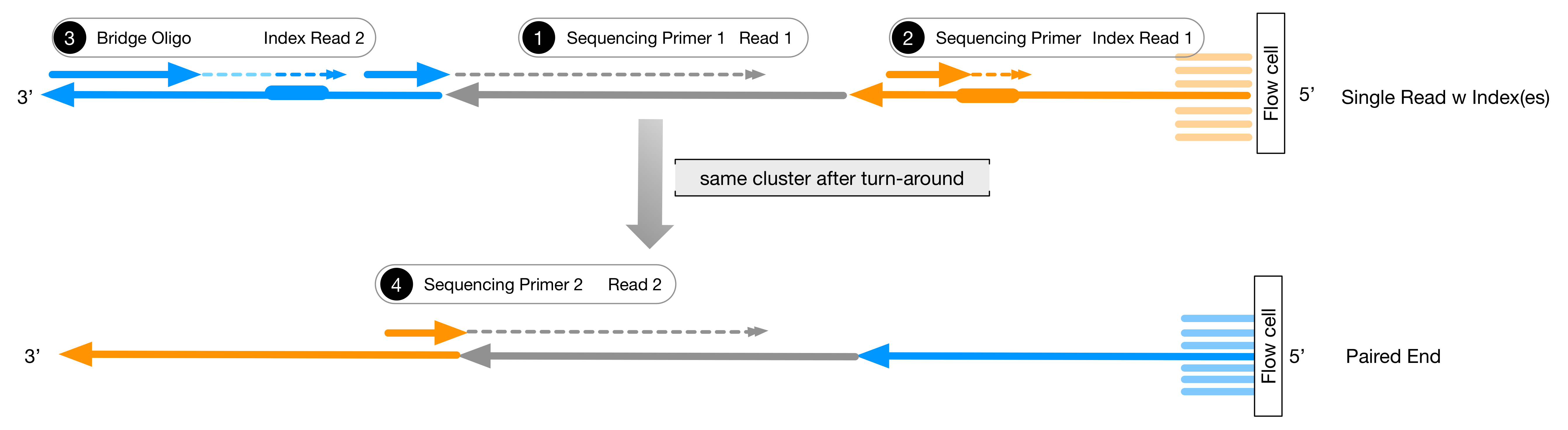
Major short-read and long-read sequencing technologies. (A) Illumina... | Download Scientific Diagram